

the ultimate power behind cancer testing.
genomic and epigenomic insights for the most complete view of cancer.1
Unlock epigenomic biomarkers
for an additional layer of information.1

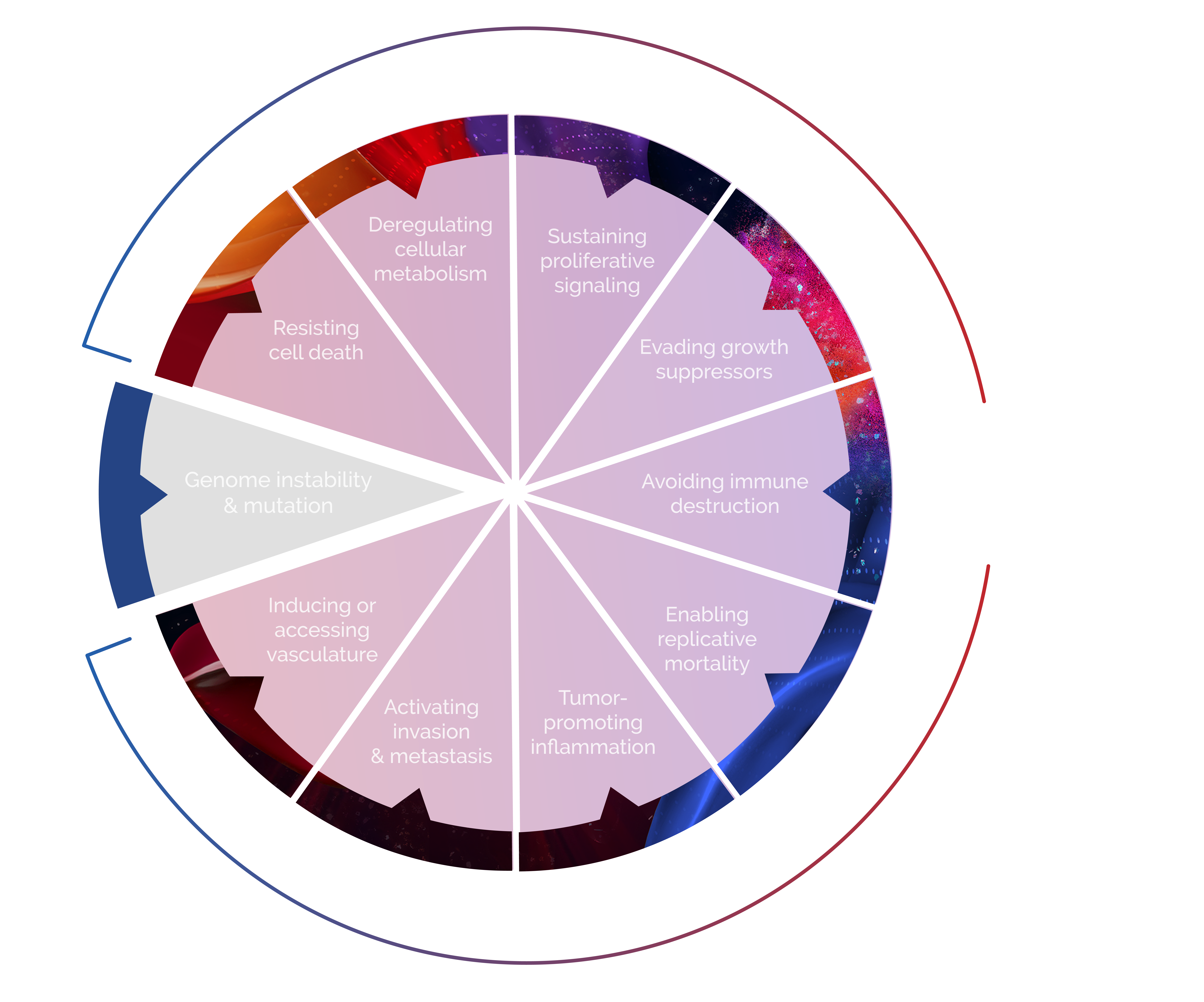
All cells in the human body share one genome, yet certain cells behave
differently, showing that genomics is only one part of a much larger picture.2-5
While analyzing the genome can provide a foundational understanding of
cancer biology, a novel approach that goes beyond genomics is needed to
uncover potential insights that may guide future breakthroughs.6

in helping us better understand
what drives cancer behavior.4
Epigenomics examines chemical and structural modifications to DNA and
histones that affect gene expression and regulate biological processes—
without altering the underlying DNA sequence.7

Genomic alterations change the underlying DNA.8


DNA sequence but can influence cancer behavior.3,7,8

examines DNA methylation—
a powerful epigenomic biomarker.1,3
Compared with normal cells, cancer cells exhibit a distinct
methylation pattern.9
Guardant Infinity maps the unique epigenomic fingerprint of cancer
by examining differences in methylation patterns.1,4

Constitutively methylated regions are segments of DNA with consistent
methylation unrelated to cancer that serve as a control.11
Unlock epigenomic biomarkers for an additional layer of information.1

All cells in the human body share one genome, yet certain cells behave differently, showing that genomics is only one part of a much larger picture.2-5

While analyzing the genome can provide a foundational understanding of cancer biology, a novel approach that goes beyond genomics is needed to uncover potential insights that may guide future breakthroughs.6

Epigenomics examines chemical and structural modifications to DNA and histones that affect gene expression and regulate biological processes—without altering the underlying DNA sequence.7

Genomic alterations change
the underlying DNA.8

Epigenomic alterations
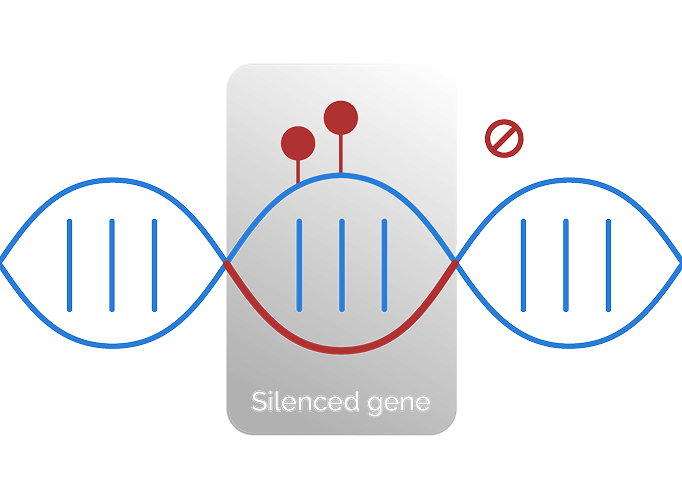
Epigenomic alterations do not change the underlying DNA sequence but can influence cancer behavior.3,7,8

Compared with normal cells, cancer cells exhibit a distinct methylation pattern.9

Constitutively methylated regions are segments of DNA with consistent
methylation unrelated to cancer that serve as a control.11

Differentially methylated regions are segments of DNA unique to cancer used for ctDNA detection and clinical insights.12
Analyze a larger set of signals to maximize sensitivity.1
Leverage new test features and applications.1

References: 1. Data on file. Guardant Health, Inc. Redwood City, CA. 2. National Human Genome Research Institute. A Brief Guide to Genomics. Published August 27, 2022. Accessed March 19, 2025. https://www.genome.gov/about-genomics/fact-sheets/A-Brief-Guide-to-Genomics 3. Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358(11):1148-1159. doi:10.1056/NEJMra072067. 4. LaFave LM, Savage RE, Buenrostro JD. Single-cell epigenomics reveals mechanisms of cancer progression. Annu Rev Cancer Biol. 2022;6:167-185. doi:10.1146/annurev-cancerbio-070620-094453. 5. Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12(1):31-46. doi:10.1158/2159-8290.CD-21-1059 6. Angeles AK, Janke F, Bauer S, Christopoulos P, Riediger AL, Sültmann H. Liquid biopsies beyond mutation calling: genomic and epigenomic features of cell-free DNA in cancer. Cancers (Basel). 2021;13(22):5615. doi:10.3390/cancers13225615 7. Al Aboud NM, Tupper C, Jialal I. Genetics, Epigenetic Mechanism. In: StatPearls. Treasure Island (FL): StatPearls Publishing; August 14, 2023. 8. Chakravarthi BV, Nepal S, Varambally S. Genomic and epigenomic alterations in cancer. Am J Pathol. 2016;186(7):1724-1735. doi:10.1016/j.ajpath.2016.02.023 9. Liu P, Zhang J, Du D, et al. Altered DNA methylation pattern reveals epigenetic regulation of Hox genes in thoracic aortic dissection and serves as a biomarker in disease diagnosis. Clin Epigenetics. 2021;13(1):124. doi:10.1186/s13148-021-01110-9 10. Cho JW, Shim HS, Lee CY, et al. The importance of enhancer methylation for epigenetic regulation of tumorigenesis in squamous lung cancer. Exp Mol Med. 2022;54(1):12-22. doi:10.1038/s12276-021-00718-4 11. Gu J, Stevens M, Xing X, et al. Mapping of variable DNA methylation across multiple cell types defines a dynamic regulatory landscape of the human genome. G3 (Bethesda). 2016;6(4):973-986. doi:10.1534/g3.115.025437 12. Peters TJ, Buckley MJ, Chen Y, Smyth GK, Goodnow CC, Clark SJ. Calling differentially methylated regions from whole genome bisulphite sequencing with DMRcate. Nucleic Acids Res. 2021;49(19):e109. doi:10.1093/nar/gkab637 13. GuardantINFINITY™ Specification Sheet. Guardant Health, Inc. Redwood City, CA.